GSE20592
|
| What
We Learned |
- These small RNAs (~18-30 nucleotides in length) function by guiding sequence-specific gene silencing at the transcriptional and/or posttranscriptional level and have been shown to play important regulatory roles in diverse biological processes. Among the small RNA classes, microRNA
(miRNA) is the most abundant class in mammals.
|
- Although sequence count data is analogous in some ways
to microarray data, the two data types differ in numerous ways.
- First, microarray data provides an analogue measure
of sequence prevalence while sequencing is inherently digital.
- Second, microarray analyses generally operate above a low background level of non-specific and
off-target probe-array binding that can complicate the analysis of low-abundance molecular species (particular
in cases where a related highly abundant product is present).
- With large enough sample sets, sequence-based analysis can avoid these background problems, allowing
exquisite sensitivity.
- Microarray and sequence-counting based approaches share certain
challenges, including biological and non-biological contamination and sample
quality and reliability.
|
- Sequencing-based miRNA profiling does not provide
absolute measurements of miRNA expression, but rather the relative counts of different miRNAs within each sample.
- miRNA-seq data are typically characterized by variances in total
counts for each sample. These, as well as sequence counts for individual miRNAs, will be subject to
large sampling noise. Moreover, in contrast to microarray data, the
miRNA-seq data involve non-negative counts.
|
- One aspect of miRNA diversity comes from the ability to
produce two distinct miRNAs (termed miR-X and miRX*) from a given hairpin precursor RNA.
- In this case, the two RNAs are distinct products of the same initial processing
product (pre-RNA), one located 5' and one 3' on this precursor.
- The standard nomenclature for miRNAs assigns the
asterisk to the less abundant of the two forms found in the first identifying study.
- Although these antisense miRNAs are low in abundance
(<100 copies in all libraries), their low concentration does not rule out their
possible biological relevance.
|
|
| What
We Did |
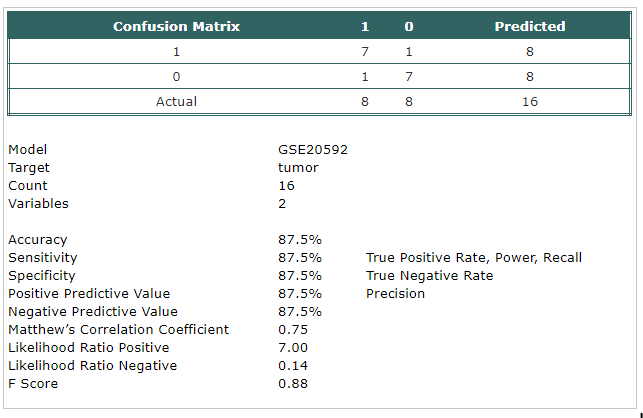
- A classification model has been built using Trainset.
The selected probes were:
- mir-125a-5p
- candidate-12-3p
|
- The model has been tested using Testset.
|
 |
|

