GSE31568
|
| What
We Learned |
- The aim of our multicenter study was to elucidate and compare blood expression profiles
of 863 miRNAs by array analysis of 454 blood samples from human individuals
for different human diseases to test for disease-specific alterations.
- Lung cancer
- Prostate
cancer (23 cases)
- Pancreatic ductal
adenocarcinoma
- Melanoma
- Ovarian
cancer
- Tumor of stomach
- Wilms tumor
- Pancreatic
tumors
- Multiple
sclerosis
- Chronic
obstructive pulmonary disease (COPD)
- Sarcoidosis
- Periodontitis
- Pancreatitis
- Acute
myocardial infarction
- Unaffected individuals or controls (70
cases)
|
- To test for technical variance, the measurements on four samples (two blood samples and two tissue
samples) have been repeated and found a median correlation of 0.97.
- The correlation between different samples was significantly lower as shown
by two-tailed unpaired Wilcoxon Mann-Whitney test (P < 0.05).
- To estimate the biological variance, blood samples taken from a healthy individual
have been analyzed at three different time points during the day (9 a.m., 12 noon and 3 p.m.),
with duplicate measurements at each time. Median correlation between the time points was 0.98 and between duplicates it was
0.99.
|
|
| What
We Did |
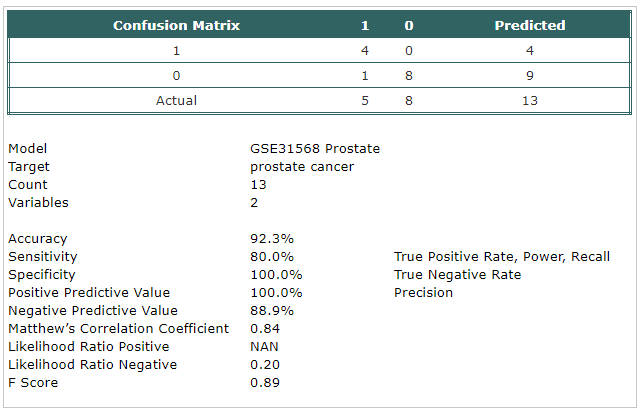
- A classification model has been built using Trainset.
The selected probes were:
- hsa-mir-519b-5p
- hsa-mir-1283
|
- The model has been tested using Testset.
|
 |
|

