GSE56350
|
| What
We Learned |
- The aim of the study was to identify a microRNA signature for metastatic CRC
(colorectal cancer) that could predict and differentiate metastatic target organ
localization.
|
- Hsa-miRNA-21, Hsa-miRNA-93, and
Hsa-miRNA-103 upregulation together with hsa-miR-566 downregulation
defined the CRC metastatic signature.
|
- This study provided the first microRNAs signature that could discriminate between
colorectal recurrences to lymph nodes and liver
and between colorectal liver metastasis and primary hepatic tumor.
|
- MicroRNAs signatures
are highly tissue specific and can be used to classify cancers, and to identify the primary tumor of a metastatic lesion of unknown
origin.
|
|
| What
We Did |
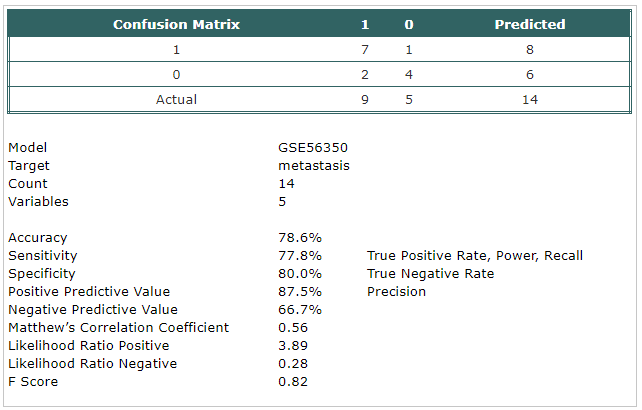
- A classification model ( metastasis tissue
against primary tumor) has been built using Trainset.
The selected probes were:
- hsa-mir-150
- hsa-mir-143
- hsa-mir-128a
- hsa-mir-122
- hsa-mir-345
|
- The model has been tested using Testset.
|
 |
|

