GSE73002
|
| What
We Learned |
- In the present study, the expression profiles of
serum miRNA in several large cohorts have been examined to identify novel
miRNA that can be used to detect early stage breast cancer.
|
- A total of 1280 serum samples of breast cancer patients stored in the
National Cancer Center Biobank were used. In addition, 2836 serum samples were
obtained from non-cancer controls, 451 from patients with other types of cancers,
and 63 from patients with non-breast benign diseases.
|
- miRNA expressions were compared between patients with breast cancer and
non-breast cancer, and a combination of five miRNA was found to be able to detect breast
cancer.
- miR-1246
- miR-1307-3p
- miR-4634
- miR-6861-5p
- miR-6875-5p
- This combination had a sensitivity of
97.3%, specificity of 82.9% and accuracy of 89.7% for breast cancer in the test cohort.
|
- A total of 400 patients with other cancers (100, 98, 50, 50, 50 and 52 cases of pancreatic, biliary
tract, colon, gastric, esophageal and hepatic cancers, respectively) and 21 patients with non-malignant pancreatic ⁄
biliary tract disease who were registered in the National Cancer Center
(NCC) Biobank were included to determine the circulating miRNA in control subjects.
- Whole miRNA expression profiles of these serum samples are present in the Gene Expression
Omnibus (GEO) database (GSE59856), and the registered data and clinical information(21) were used in the present study.
|
- To normalize the signals across the different
microarrays tested, three pre-selected internal control miRNA (miR-149-3p, miR-2861 and miR-4463), which had been
stably detected in more than 500 serum samples, were used.
- Each miRNA signal value was standardized with the ratio
of the average signal value of the three internal control miRNA to the pre-set value.
|
- The diagnostic index showed high performance for
all cancer stages (stage 0, 98.0%; stage 1, 98.1%; stage 2, 95.7%;
stage 3, 100%; stage 4, 96.2%), and TNM grades (T: Tis, 97.7%; T1, 98.2%; T2, 95.8%; T3, 90.9%; T4, 97.1%; N: N0,
97.2%; N1, 98.2%; N2, 92.9%; N3, 100%; and M: M0, 97.3%; M1, 96.2%).
|
- A problem with the developed diagnostic index is that it
cannot distinguish benign breast diseases from breast
cancer.
- With this diagnostic index, benign breast diseases were identified
as breast cancer. Therefore, it might be used for breast hyperplasia.
- However, as one aim of the present study was to
identify an initial screening technique that provides good reasoning
and motivation for more women to undergo mammography, we believe that the problem of
false-positive results of the diagnostic index caused by benign breast diseases is
acceptable at the present time.
- However, studies should be performed in the future to
improve the diagnostic index so that it can distinguish benign breast diseases from breast cancer.
|
|
| What
We Did |
- Here, we compared breast benign disease cases
with non-cancer cases.
|
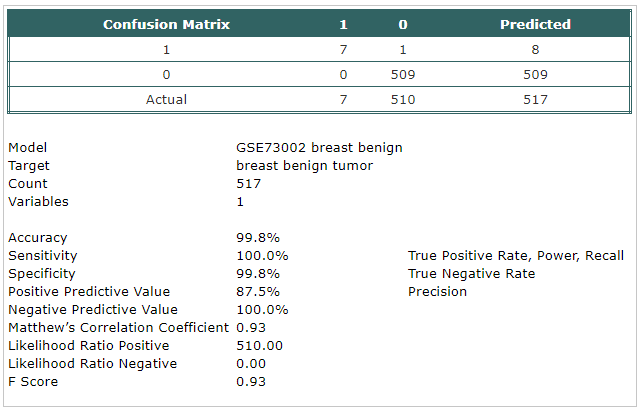
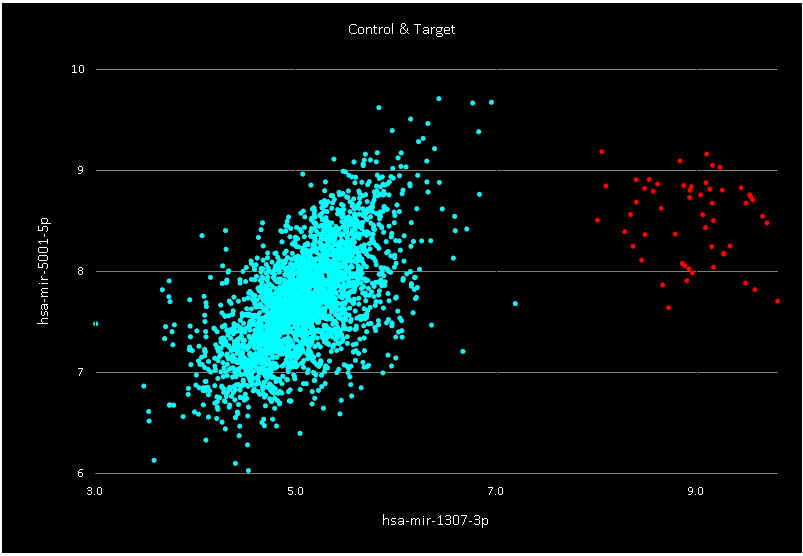
- A classification model has been built using Trainset.
The selected probe was:
- hsa-mir-1307-3p
|
- The model has been tested using Testset.
|
 |
|
 |
|



